Dataset |
n
| Prevalence | %t
| Full dataset accuracy | Optimal vs. 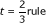
| Optimal vs. 
|
|---|
Rosenwald | 240 | 52% | 63% | 0.96 | 0.001 | 0.002 |
Boer | 152 | 53% | 53% | 0.98 | 0.004 | 2e-4 |
Golub | 72 | 65% | 56% | 0.95 | 0.002 | 0.004 |
Sun | 131 | 62% | 31% | 0.83 | 0.022 | 0.008 |
van't Veer | 117 | 67% | 26% | 0.78 | 0.004 | 0.001 |
- Nonparametric bootstrap with smooth spline (or isotonic regression) learning curve method results [Additional file 1]. n is the total number of samples from the two classes, and "Prevalence" is the prevalence of the majority class. %t is the percent of samples allocated to the training set under optimal allocation, t/n ·100%. "Full dataset accuracy" is the estimated mean accuracy on the full dataset of size n. "Optimal vs.
 rule" is the difference between the root mean squared error for an optimal training set allocation and for the "2/3 rds to training set" allocation rule. The rightmost column is for the "1/2 to training set" allocation rule. Classes for datasets are: Germinal Center B-cell-like lymphoma versus other (Rosenwald et al., 2002), renal clear cell carcinoma primary tumor versus control normal kidney tissue (Boer et al., 2001), acute myelogenous leukemia versus acute lymphoblastic leukemia (Golub et al., 1999), glioblastoma versus oligodendroglioma (Sun et al., 2006), grade 1/2 versus grade 3 lung cancer (van't Veer et al., 2002).
rule" is the difference between the root mean squared error for an optimal training set allocation and for the "2/3 rds to training set" allocation rule. The rightmost column is for the "1/2 to training set" allocation rule. Classes for datasets are: Germinal Center B-cell-like lymphoma versus other (Rosenwald et al., 2002), renal clear cell carcinoma primary tumor versus control normal kidney tissue (Boer et al., 2001), acute myelogenous leukemia versus acute lymphoblastic leukemia (Golub et al., 1999), glioblastoma versus oligodendroglioma (Sun et al., 2006), grade 1/2 versus grade 3 lung cancer (van't Veer et al., 2002).
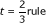

 rule" is the difference between the root mean squared error for an optimal training set allocation and for the "2/3 rds to training set" allocation rule. The rightmost column is for the "1/2 to training set" allocation rule. Classes for datasets are: Germinal Center B-cell-like lymphoma versus other (Rosenwald et al., 2002), renal clear cell carcinoma primary tumor versus control normal kidney tissue (Boer et al., 2001), acute myelogenous leukemia versus acute lymphoblastic leukemia (Golub et al., 1999), glioblastoma versus oligodendroglioma (Sun et al., 2006), grade 1/2 versus grade 3 lung cancer (van't Veer et al., 2002).
rule" is the difference between the root mean squared error for an optimal training set allocation and for the "2/3 rds to training set" allocation rule. The rightmost column is for the "1/2 to training set" allocation rule. Classes for datasets are: Germinal Center B-cell-like lymphoma versus other (Rosenwald et al., 2002), renal clear cell carcinoma primary tumor versus control normal kidney tissue (Boer et al., 2001), acute myelogenous leukemia versus acute lymphoblastic leukemia (Golub et al., 1999), glioblastoma versus oligodendroglioma (Sun et al., 2006), grade 1/2 versus grade 3 lung cancer (van't Veer et al., 2002).